This post was co-written with Rajiv Arora, Director of Data Science Platform at Gilead Life Sciences.
Gilead Sciences, Inc. is a biopharmaceutical company committed to advancing innovative medicines to prevent and treat life-threatening diseases, including HIV, viral hepatitis, inflammation, and cancer. A leader in virology, Gilead historically relied on these drugs for growth but now through strategic investments, Gilead is expanding and increasing their focus in oncology, having acquired Kite and Immunomedics to boost their exposure to cell therapy and non-cell therapy, making it the primary growth engine. Because Gilead is expanding into biologics and large molecule therapies, and has an ambitious goal of launching 10 innovative therapies by 2030, there is heavy emphasis on using data with AI and machine learning (ML) to accelerate the drug discovery pipeline.
Amazon Redshift Serverless is a fully managed cloud data warehouse that allows you to seamlessly create your data warehouse with no infrastructure management required. You pay only for the compute resources and storage that you use. Redshift Serverless measures data warehouse capacity in Redshift Processing Units (RPUs), which are part of the compute resources. All of the data stored in your warehouse, such as tables, views, and users, make up a namespace in Redshift Serverless.
One of the benefits of Redshift Serverless is that you don’t need to size your data warehouse for your peak workload. The peak workload includes loading periodic large datasets in multi-terabyte range. You can set a base RPU from 8 up to 512 and Redshift Serverless will automatically scale the RPUs to meet your workload demands. This makes it straightforward to manage your data warehouse in a cost-effective manner.
In this post, we share how Gilead collaborated with AWS to redesign their data ingestion process. They used Redshift Serverless as their data producer to load third-party medical claims data in a fast and cost-effective way, reducing load times from days to hours.
Gilead use case
Gilead loads a variety of data from hundreds of sources to their R&D data environment. They recently needed to do a monthly load of 140 TB of uncompressed healthcare claims data in under 24 hours after receiving it to provide analysts and data scientists with up-to-date information on a patient’s healthcare journey. This data volume is expected to increase monthly and is fully refreshed each month. The 3-node RA3 16XL provisioned cluster that had previously been hosting their warehouse was taking around 12 hours to ingest this data to Amazon Redshift, and Gilead was looking to optimize the data ingestion process in a more dynamic manner. Working with Amazon Redshift specialists from AWS, Gilead chose Redshift Serverless as a way to cost-effectively load this data and then use Redshift data sharing to share the final dataset to two additional Redshift data warehouses for end-user queries.
Loading data is a key process for any analytical system, including Amazon Redshift. When loading very large datasets, it’s important to not only load the data as quickly as possible but also in a way that optimizes the consumption queries.
Gilead’s healthcare claims data took 40 hours to load, which meant delays in using the data for downstream processes. The teams sought improvements, targeting a maximum 24-hour SLA for the load. They achieved the load in 8 hours, an 80% reduction in time to make data available.
Solution overview
After collaborating, the Gilead and AWS teams decided on a two-step process to load the data to Amazon Redshift. First, the data was loaded without a distkey and sortkey, which let the load process use the full parallel resources of the cluster. Then we used a deep copy to redistribute this data and add the desired distribution and sort characteristics.
The solution uses Redshift Serverless. The team wanted to ingest data to meet the required SLA, and the following approaches were benchmarked:
- COPY command – The COPY command uses the Amazon Redshift massively parallel processing (MPP) architecture to read and load data in parallel from files on Amazon Simple Storage Service (Amazon S3)
- Data lake analytics – Amazon Redshift Spectrum is used to query data directly from files on Amazon S3 by selecting a subset of columns and avoiding the intermediate step of copying data to staging table
Initial Solution approach: Single COPY command
The team determined it would be more effective to apply the distribution and sort keys in a post-copy step. The data was loaded first using automatic distribution of data. This took roughly 12 hours to complete. The team created open and closed claims tables with defined dist keys and with 20% of the columns to alleviate the need to query the larger table. With this success, we learned that we can still improve the big copy, as detailed in the following sections.
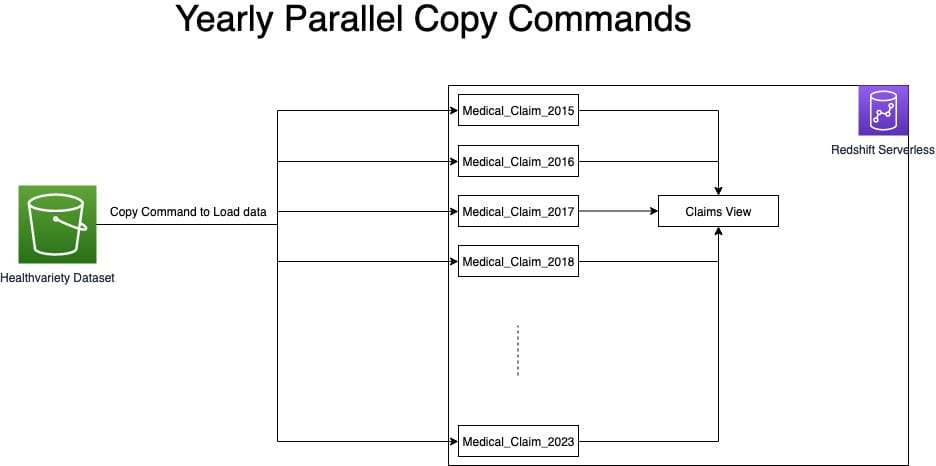
Proposed Solution approach 1: Parallel COPY command
Based on the initial solution approach above, the team tested yearly parallel copy commands as illustrated in the following diagram.
Below are the findings and learnings from this approach:
- Ingesting data for 4 years using parallel copy showed a 25% performance improvement over the single copy command.
- Compared to Initial solution approach, where we were taking 12 hours to ingest the data, we further optimized this runtime by 67% by segregating the data ingestion into separate yearly staging tables and running parallel copy commands.
- After the data was loaded into staging yearly tables, we created the open and closed claim tables with an auto
distkeywith the subset of columns required for larger reporting groups. It took an additional 1 hour to create.
The team used a manifest file to make sure that the COPY command loads all of the required files for the respective year for ingesting.
Proposed Solution approach 2: Data Lake analytics
The team used this approach with Redshift Spectrum to load only the required columns to Redshift Serverless, which avoided loading data into multiple yearly tables and directly to a single table. The following diagram illustrates this approach.
The workflow consists of the following steps:
- Crawl the files using AWS Glue.
- Create a data lake external schema and table in Redshift Serverless.
- Create two separate claims table for open and closed claims because open claims are most frequently consumed and are 20% of the columns and 100% of the data.
- Create open and closed tables with selective columns needed for optimal performance optimization during consumption instead of all columns in the original third-party dataset. The data volume distribution is as follows:
- Total number of open claims records = 50 billion
- Total number of closed claims records = 200 billion
- Overall, total number of records = 250 billion
- Distribute open and closed tables with a customer-identified
distkey. - Configure data ingestion into open and closed claims tables combined using Redshift Serverless with 512 RPUs. This took 1.5 hours, which is further improved by 70% compared to scenario 1. We chose 512 RPUs in order to load data in the fastest way possible.
In this method, data ingestion was streamlined by only loading essential fields from the medical claims dataset and by splitting the table into open and closed claims. Open claims data is most frequently accessed and constitutes only 20% of columns so by splitting the tables. The team not only improved the ingestion performance but also consumption.
Amazon Redshift recently launched automatic mounting of AWS Glue Data Catalog, making it easier to run data lake analytics without manually creating external schemas. You can query data lake tables directly from Amazon Redshift Query Editor v2 or your favorite SQL editors.
Recommendations and best practices
Consider the following recommendations when loading large-scale data in Amazon Redshift.
- Use Redshift Serverless with maximum 512 RPUs to efficiently and quickly load data
- Depending on consumption use case and query pattern, adopt either of the following approaches:
- When consumption queries require only selected fields from the dataset and most frequently access a subset of data, use data lake queries to load only the relevant columns from Amazon S3 into Amazon Redshift
- When consumption queries require all fields, use COPY commands with a manifest file to ingest data in parallel into multiple logically separated tables and create a database view with UNION ALL of all tables
- Avoid using varchar(max) while creating tables and create VARCHAR columns with the right size
Final Architecture
The following diagram shows the high-level final architecture that was implemented.
Conclusion
With the scalability of Redshift Serverless, data sharing to decouple ingestion from consumption workloads, and data lake analytics to ingest data, Gilead made their 140 TB dataset available to their analysts within hours of it being delivered. The innovative architecture of using a serverless ingestion data warehouse, a serverless consumption data warehouse for power users, and their original 3-node provisioned cluster for standard queries gives Gilead isolation to ensure data loads don’t affect their users. The architecture provides scalability to serve infrequent large queries with their serverless consumer along with the benefit of a fixed-cost and fixed-performance option of their provisioned cluster for their standard user queries. Due to the monthly schedule of the data load and the variable need for large queries by consumers, Redshift Serverless proved to be a cost-effective option compared to simply increasing the provisioned cluster to serve each of these use cases.
This split producer/consumer model of using Redshift serverless can bring benefits to many workloads that have similar performance characteristics to Gilead’s warehouse. Customers regularly run large data loads infrequently, and those processes compete with user queries. With this pattern, you can rely on your queries to perform consistently regardless of whether new data is being loaded to the system. This strikes a balance between minimizing cost while maintaining performance and frees the system administrators to load data without affecting users.
About the Authors
 Rajiv Arora is a Director of Clinical Data Science at Gilead Sciences with over 20 years of experience in the industry. He is responsible for the multi-modal data platform for the development organization and supports all statistical and predictive analytical infrastructure for RWE and Advanced Analytical functions.
Rajiv Arora is a Director of Clinical Data Science at Gilead Sciences with over 20 years of experience in the industry. He is responsible for the multi-modal data platform for the development organization and supports all statistical and predictive analytical infrastructure for RWE and Advanced Analytical functions.
 Ritesh Kumar Sinha is an Analytics Specialist Solutions Architect based out of San Francisco. He has helped customers build scalable data warehousing and big data solutions for over 16 years. He loves to design and build efficient end-to-end solutions on AWS. In his spare time, he loves reading, walking, and doing yoga.
Ritesh Kumar Sinha is an Analytics Specialist Solutions Architect based out of San Francisco. He has helped customers build scalable data warehousing and big data solutions for over 16 years. He loves to design and build efficient end-to-end solutions on AWS. In his spare time, he loves reading, walking, and doing yoga.
 Raks Khare is an Analytics Specialist Solutions Architect at AWS based out of Pennsylvania. He helps customers architect data analytics solutions at scale on the AWS platform.
Raks Khare is an Analytics Specialist Solutions Architect at AWS based out of Pennsylvania. He helps customers architect data analytics solutions at scale on the AWS platform.
 Brent Strong is a Senior Solutions Architect in the Healthcare and Life Sciences team at AWS. He has more than 15 years of experience in the industry, focusing on data and analytics and DevOps. At AWS, he works closely with large Life Sciences customers to help them deliver new and innovative treatments.
Brent Strong is a Senior Solutions Architect in the Healthcare and Life Sciences team at AWS. He has more than 15 years of experience in the industry, focusing on data and analytics and DevOps. At AWS, he works closely with large Life Sciences customers to help them deliver new and innovative treatments.
 Phil Bates is a Senior Analytics Specialist Solutions Architect at AWS with over 25 years of data warehouse experience.
Phil Bates is a Senior Analytics Specialist Solutions Architect at AWS with over 25 years of data warehouse experience.